KEGG
Outline
- Fetch Separate KEGG Pathways
- Fetch and Merge Pathways
- Clean up or subset fetched KEGG network
- Overlay microarray expression onto KEGG network
- Overlay metabolomics data onto KEGG network (Not done yet)
Fetch Separate KEGG Pathway
Open Mango and click on Databases/KEGG in the menu.
A dialog window will pop and attempt to connect to KEGG database. A list of the current organisms on KEGG will be fetched. This may take up to a minute depending on your internet speed.
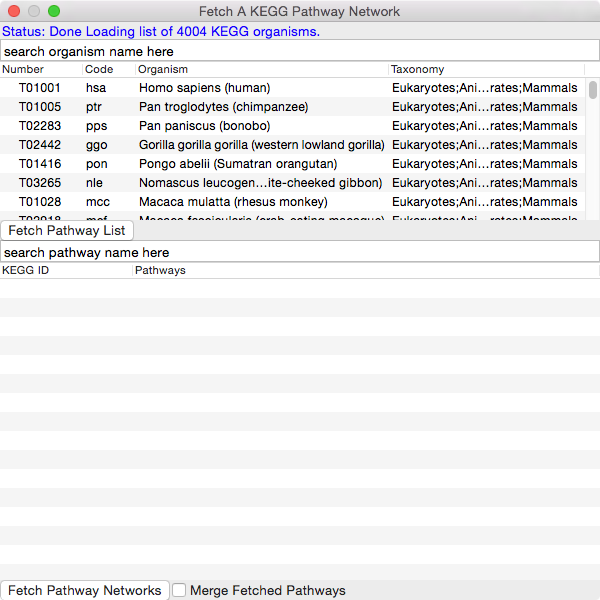
Select an organism from the list so it's highlighted and then click the Fetch Pathway List button.
You may also type into the search box and press Enter/Return to search for a particular organism. Pressing Enter multiple times will search for the next match. Clicking on the header will sort the list by column. By default, KEGG organisms are listed by taxonomic distance from human.
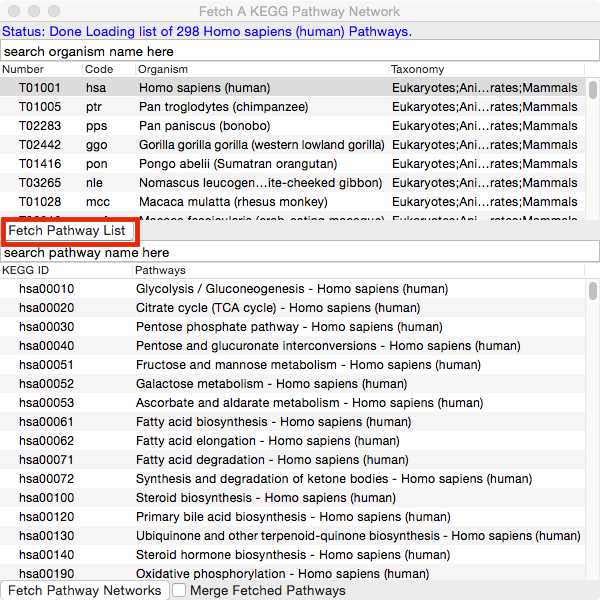
Select one pathway (click) or multiple pathways (ctrl+click, or shift+click) and hit the Fetch Pathway Networks button.
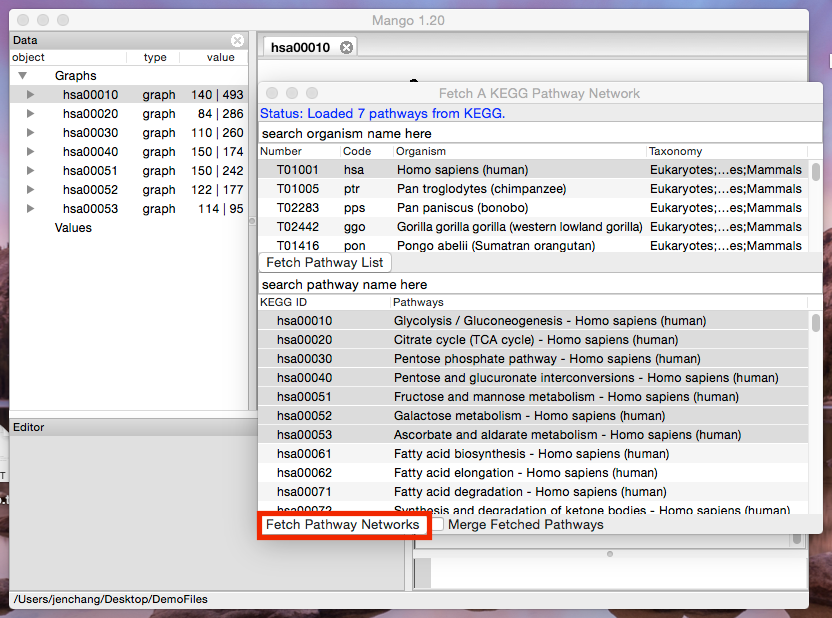
Each will be KEGG pathway will be loaded as a separate network.
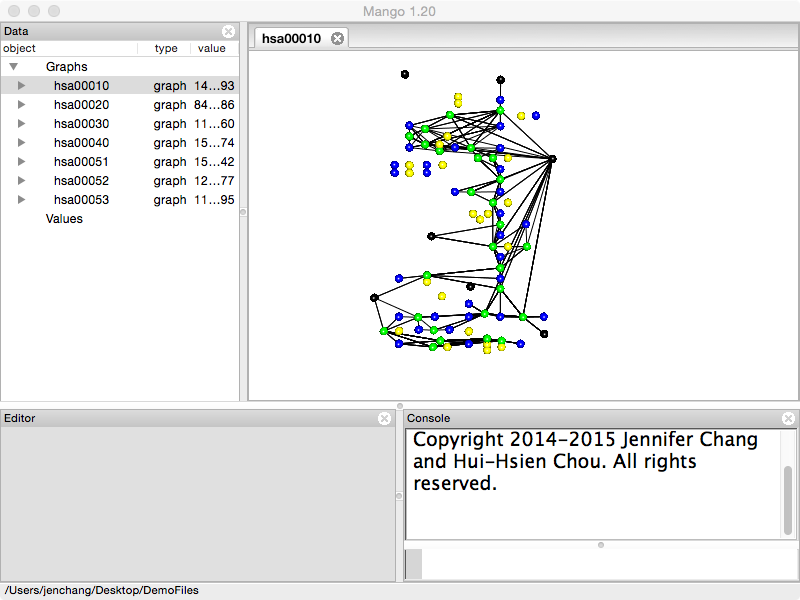
Double click the pathway and notice how it retains the original xy coordinates from KEGG. By default genes are colored green, compounds are colored blue, and orthologs (not within this species) are colored yellow.

Fetch Merged KEGG Pathways
Open the KEGG dialog window again, and this time check the box next to Merge Fetched Pathways and hit the Fetch Pathway Networks button.
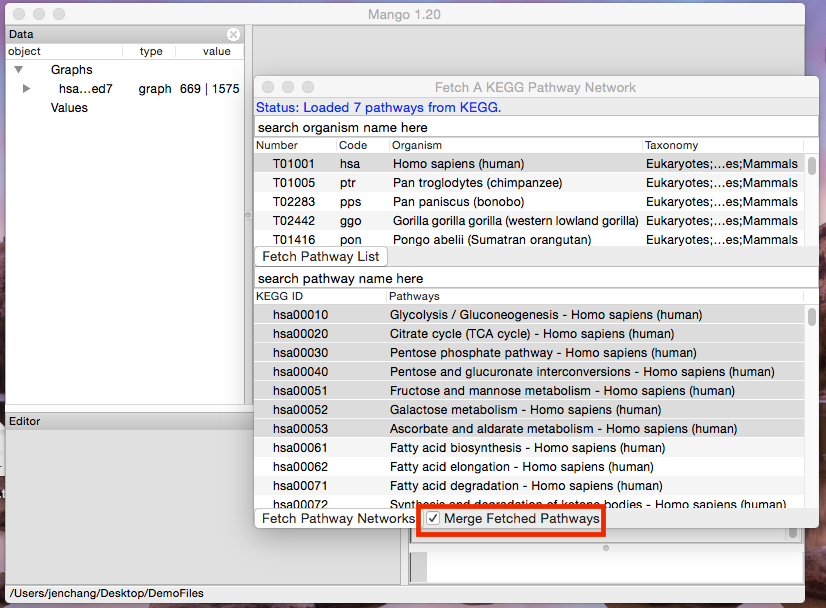
All selected pathways are merged into one network where each pathway has a different _z value.
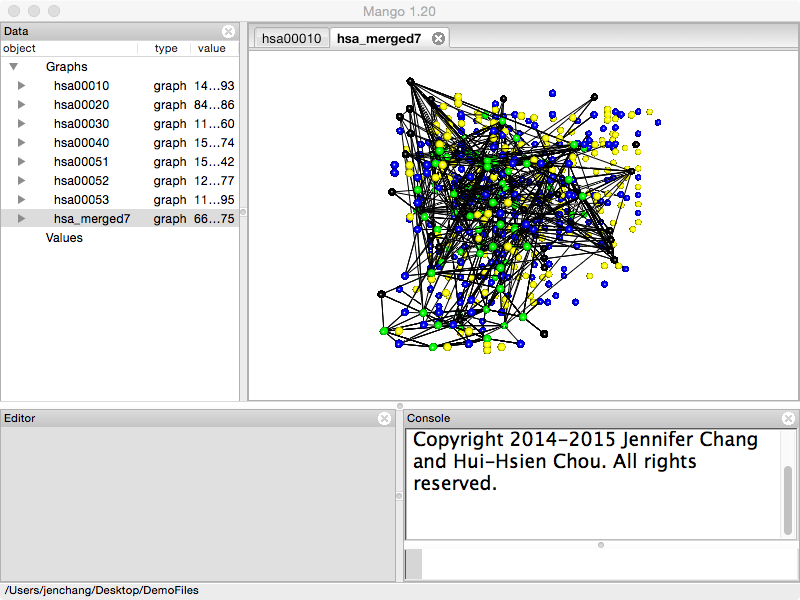
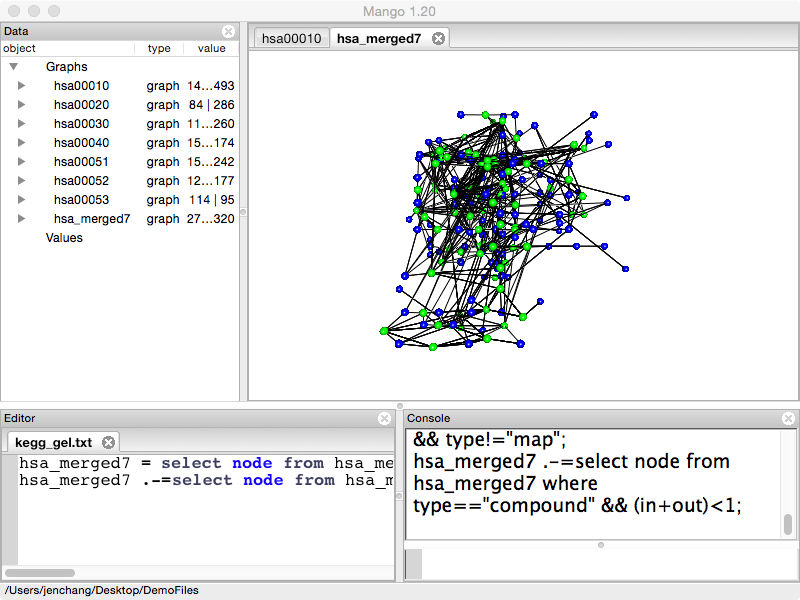
Clean Up and Subset KEGG Pathway
- Take out pathway nodes
- Remove Orthologs not in this species
- Remove compounds that are not involved in reactions in this species
We will continue to use the hsa_merged7 network for the code examples.
hsa_merged7=select node from hsa_merged7 where type!="ortholog" && type!="map";
hsa_merged7.-=select node from hsa_merged7 where type=="compound" && (in+out)<1;
Gene to Gene Network
Since KEGG contains type==ECrel or gene to gene connections as well as reactions compound to gene to compound connections.
You can get the gene to gene network using the following command:
auto ggnet = select link from hsa_merged7 where type=="ECrel";
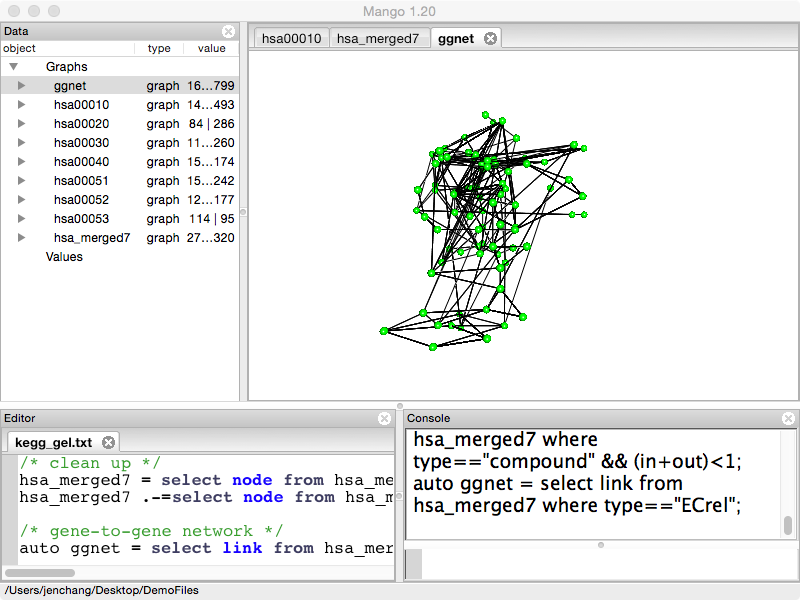
Reaction Network
Will contain compound to gene to compound links. Drop gene to gene links.
auto rxnet = select link from hsa_merged7 where type!="ECrel";
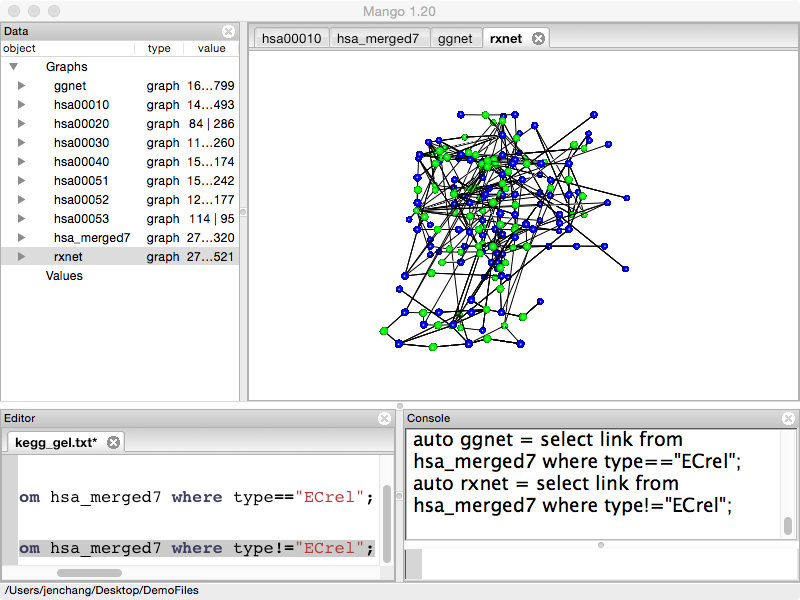
Combine with Microarray Expression Data
See Case Study 3